Our lab is also interested in the discovery, structure and mechanism of enzymes, to enable the further development and engineering to enzymes with improve properties for synthetic applications.
Amide ligases: Most recently we discovered a new family of amide ligase enzymes (CfaL), which catalyse the synthesis of amides from a large variety of different carboxylic acid and amino acid substrates [1]. Amide bonds are fundamental in nature and are present in many of leading pharmaceuticals and other valuable products. Although chemical coupling of acids and amines is relatively simple, it often requires three steps, protect–couple–deprotect, to install each amide. Stoichiometric quantities of expensive and deleterious coupling reagents are typically required and purification can be problematic. We showed that CfaL have many advantages over traditional synthetic methods and other biocatalytic approaches, providing a cleaner, more sustainable, scalable and selective (obviate the need for protecting group), recyclable, tunable and less expensive route to important amides [1].
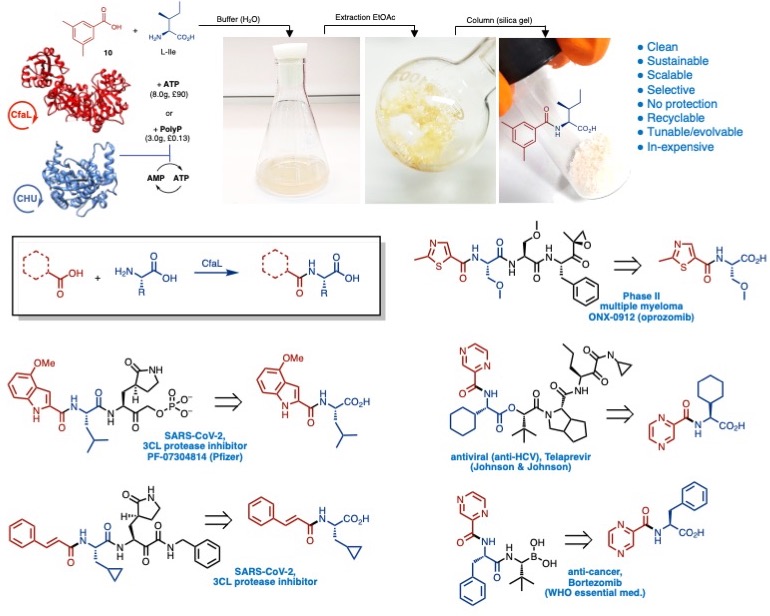
Solving the x-ray crystal structures CfaL enzymes provided valuable information which led to mutants, via structure guided mutagenesis, with improved stability and activities. We also showed how these ligases can be used for kinetic resolutions of racemic acid donor and amine acceptor substrates. Furthermore, we were able to demonstrate that these enzymes function efficiently at gram scale and preparation of precursors for important pharmaceuticals such as cancer treatments or potential COVID-19 drugs.
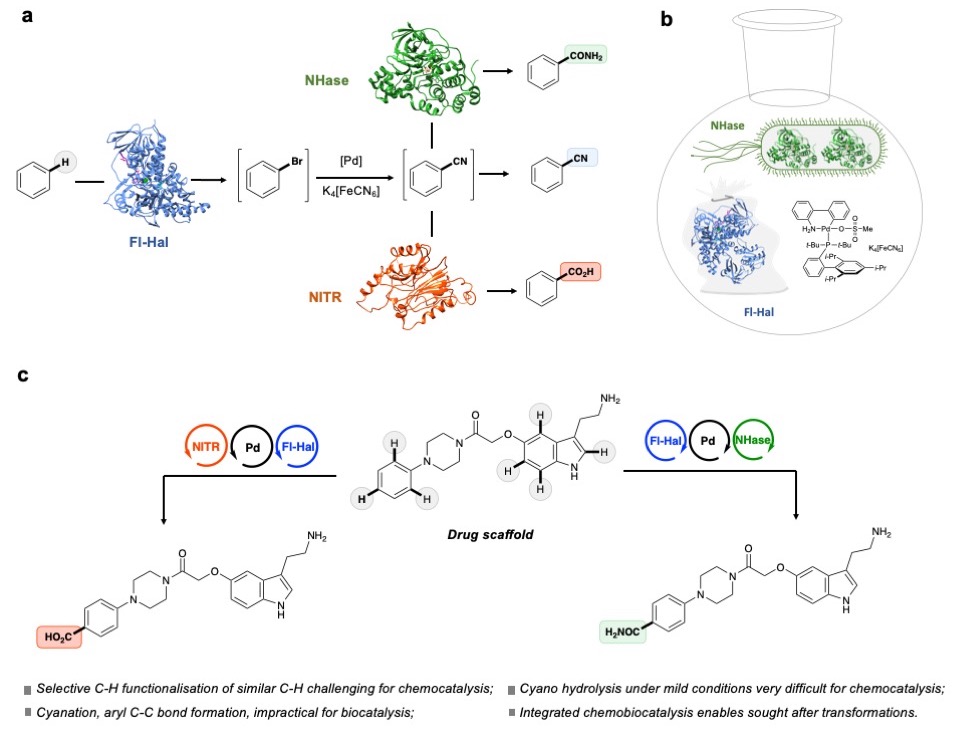
Halogenases & integrated catalysis: We have also been exploring the synthetic application of halogenase enzymes and integrated catalysis. For example, we employed structure guided mutagenesis and directed evolution to broaden the substrate scope and switch the regioselectivities of various bacterial and fungal halogenase enzymes [2, 3, 4, 15]. Our lab also demonstrated how halogenases can be integrated with palladium-catalyzed cross coupling chemistry, in one-pot reactions, to affect the direct regioselective arylation or alkenylation of C-H positions in various aromatic scaffolds [6]; such transformations are inaccessible using stand-alone chemo- or bio-catalysis. We recently extended this concept, developing programmable, regioselective C−H bond functionalization methodologies for the installation of versatile nitrile, amide and carboxylic acid moieties through integration of halogenase enzymes with palladium-catalysed cyanation and subsequent incorporation of nitrile hydratase or nitrilase enzymes [7]. Using two- or three-component chemobiocatalytic systems, the regioselective synthesis of complex target molecules, including pharmaceuticals, can be achieved in a one-pot process operable on a gram scale.
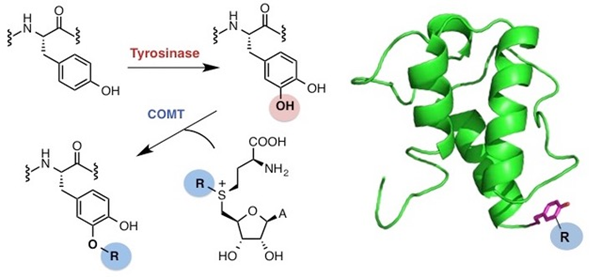
Methyltransferase & alkyl diversification: S-adenosyl methionine (SAM)-dependent methyltransferase enzymes are also of major interest. We have characterised and engineered various methyltransferases, to extend their substrate scope and to accept SAM analogues, with alternative substituents on the sulphonium centre, demonstrating how these enzymes can be used in the alkyl-diversification of bioactive scaffolds [8, 9, 10, 11, 12]. We determined the effects of active site modification and quaternary structure on the regioselectivity of catechol-O-methyltransferase (COMT), as well as developing improved COMT enzymes for the production of vanillin and ethyl vanillin [8]. An enzyme cascade including COMT and tyrosinase (an hydroxylase) was also developed that enables the selective derivatisation of tyrosine residues in peptides and proteins using SAM analogues for potential labelling, imaging and therapeutic applications [9].
Our lab also characterised the methyltransferase RapM demonstrating how it can be used for the regioselective alkylation of the important immunosuppressive agent rapamycin [10]. In addition, we obtained the first crystal structure of CNMT which, along with mutagenesis studies, defines the enzymes active site architecture [11]. CNMT was then used in the alkyl-diversification of privileged tetrahydroisoquinoline pharmacophores [12].
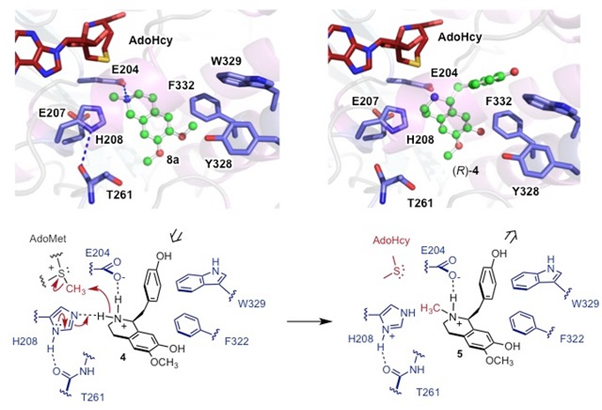
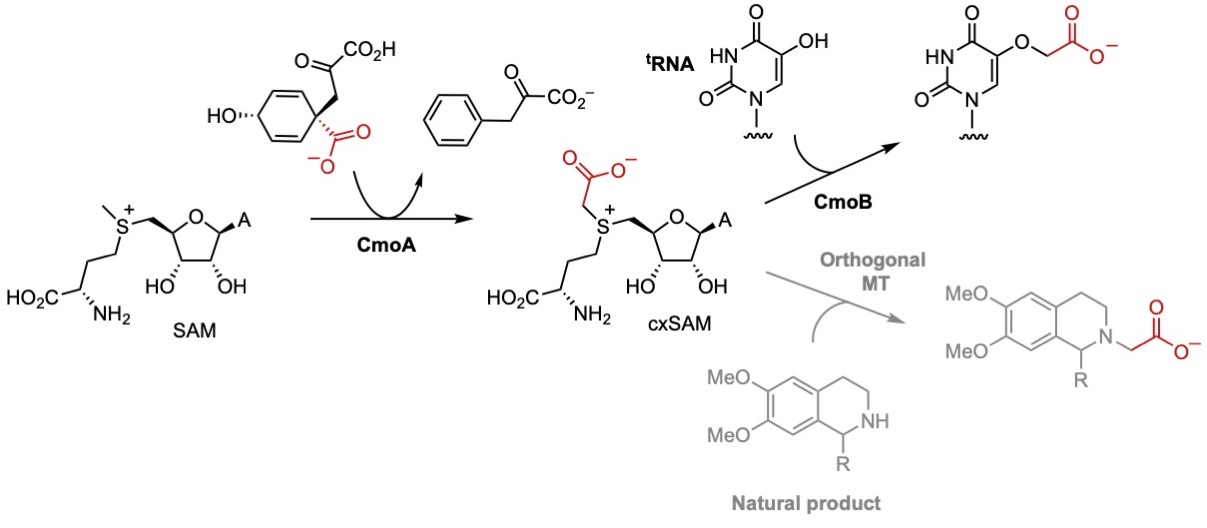
We also showed how MT enzymes can utilise a naturally occurring SAM analogues carboxy-SAM (cxSAM) which is involved in carboxylmethylation of a tRNA substrate. We used cxSAM to catalyse carboxymethylation of tetrahydroisoquinoline (THIQ) and catechol substrates using engineered CNMT and COMT enzymes. Site-directed mutagenesis was used to create orthogonal CNMT and COMT variants possessing improved catalytic activity and selectivity for cxSAM. The subsequent coupling of these orthogonal MT, with the cxSAM generating enzyme CmoA, resulted in more efficient and selective carboxymethylation pathways. An enzymatic approach was also developed to generate a previously undescribed co-factor, carboxy-S-adenosyl-l-ethionine (cxSAE), thereby enabling the stereoselective transfer of a chiral 1-carboxyethyl group to the substrate.
Other examples of enzymatic synthesis: Our team demonstrated how an engineered variant of tryptophan synthase from Salmonella (StTrpS) can catalyse the efficient condensation of L‐threonine and various indoles to generate β‐methyltryptophans and derivatives in a single step [13]. In earlier studies, we elucidated the structure, mechanism and developed directed evolution approaches to extend the biocatalytic repertoire of the aryl malonate decarboxylases [14, 15] Finally we have provided the first example of a temperature dependent switch in enzyme class, showing how phenylalanine aminomutase enzymes become lyase enzymes at elevated temperatures [16]
[1] M. Winn, F. Wang, M. Rowlinson, L. Bering, D. Francis, C. Levy &
J Micklefield Nature 2021, 593, 391–398; [2] J. Latham, E. Brandenburger, S. A. Shepherd, B.R.K. Menon & J.Micklefield. Chem. Rev. 2017, 118, 232-269; [3] R. K. Menon, E. Brandenburger, H. H. Sharif, U. Klemstein, S.A. Shepherd, M. F. Greaney & J. Micklefield. Angew. Chem. Int. Ed. 2017, 56, 11841 –11845; [4] S. A. Shepherd, C. Karthikeyan, J. Latham, A.-W. Struck, M. L. Thompson, B. Menon, M. Styles, C. Levy, D. Leys & J. Micklefield. Chemical Science2015, 6, 3454-3460; [5] S. A. Shepherd, B. R. K. Menon, H. Fisk, A.-W. Struck, C. Levy, D. Leys & J. Micklefield. ChemBioChem2016, 17, 821–824; [6] J. Latham, J.-M. Henry, H. H. Sharif, B. R. K. Menon, S. A. Shepherd, M. F. Greaney & J.Micklefield. Nature Commun.2016, 7, 11873; [7] E. J. Craven, J. Latham, S. A. Shepherd, I. Khan, A. Diaz-Rodriguez, M. F. Greaney & J. Micklefield Nature Catalysis 2021, 4, 385–394; [8] M. L. Thompson, C. Levy, S. A. Shepherd, D. Leys, and J. Micklefield Angew. Chem. Int. Ed. 2016, 55, 2683-2687; [9] A.-W. Struck, M. R. Bennett, S. A. Shepherd, B. J. C. Law, Y. Zhuo, L. S. Wong, J. Micklefield. J. Am. Chem. Soc.2016, 138, 3038-3045; [10] B. J. C. Law, A.-W. Struck, M. R. Bennett, B. Wilkinson and J. Micklefield. Chemical Science 2015, 6, 2885-2892; [11] M. R. Bennett, M. L. Thompson, S. A. Shepherd, M. S. Dunstan, A. J. Herbert, D. R. M. Smith, V. A. Cronin, B. R. K. Menon, C. Levy & J. Micklefield Angew. Chem. Int. Ed.2018, 57, 10600-10604; [12] A. J. Herbert, S. A. Shepherd, V. A. Cronin, M. R. Bennett, R. Sung & J Micklefield Angew. Chem. Int. Ed. 2020, 59, 2–9; [13] D. Francis, M. Winn, J. Latham, M. F. Greaney & J. Micklefield. ChemBioChem.2017, 18, 382-386; [14] K. Okrasa, C. Levy, M. Wilding, M. Goodall, N. Baudendistel. B. Hauer, D. Leys, J. Micklefield. Angew. Chem. Int. Ed.2009, 48, 7691-7694; [15] R. Lewin, M. Goodall, M. L.Thompson, J. Leigh, M. Breuer, K. Baldenius & J. Micklefield. Chem. Eur. J.,2015, 17, 6557-6563; [16] C. Chesters, M. Wilding, M. Goodall, J. Micklefield. Angew. Chem. Int. Ed. 2012, 51, 4344-4348.