Our lab has worked on nucleic acid chemistry and biology for many years. In fact the first work published by our lab [1, 2] focused on the chemical synthesis and evaluation of novel backbone modified nucleic acids as antisense agents and in other applications [e.g. 1, 2, 3, 4]. Currently we are developing new and more sustainable ways of producing new modified nucleic acids for range of different important therapeutics uses. Our lab also has many years of experience in the enzymatic synthesis of nucleic acids. Notably we developed the first orthogonal riboswitches and aptamers (RNA elements) that recognise small molecule ligands for in vivo applications as gene expression tools and as biosensors.
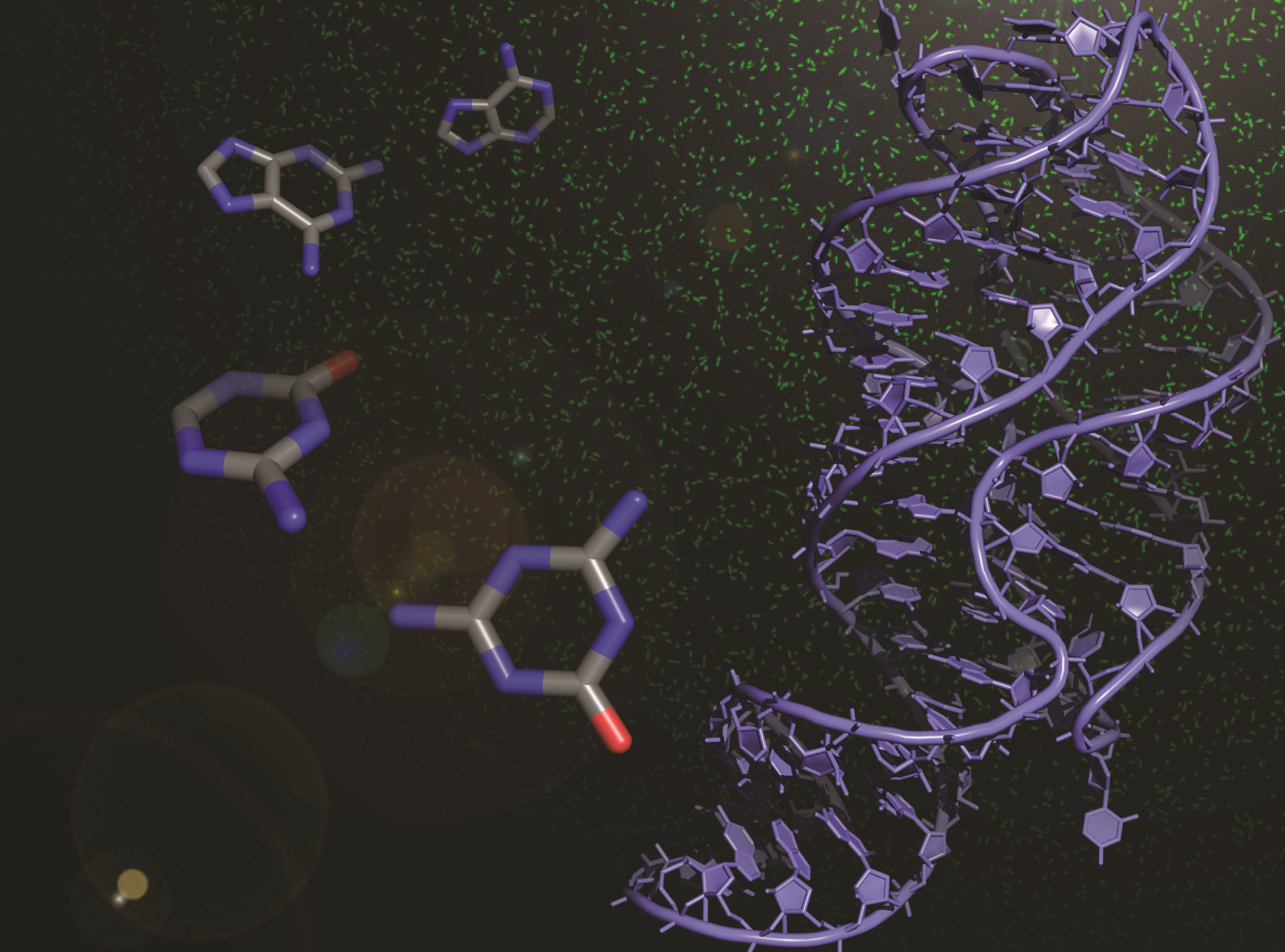
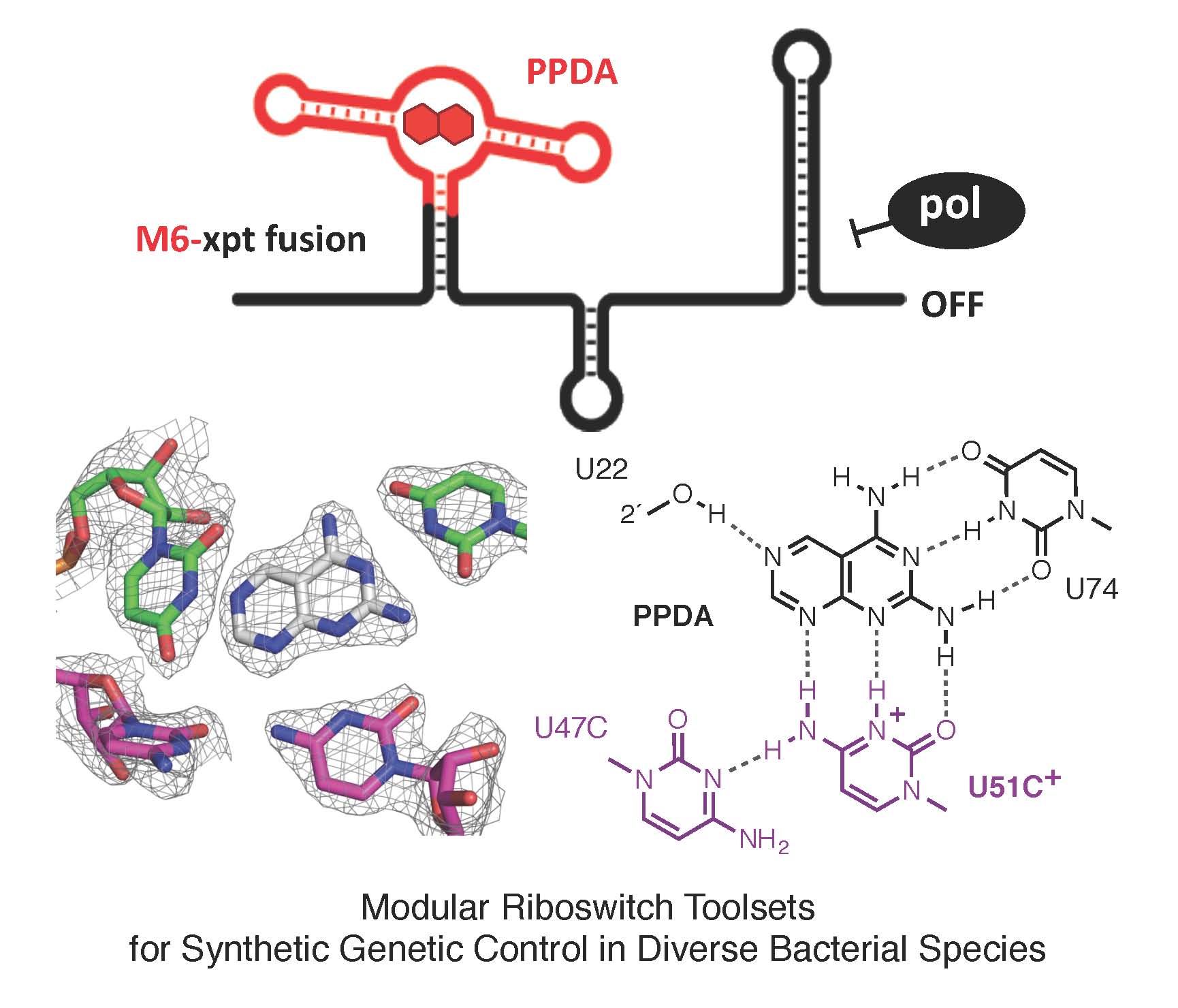
Riboswitches, present within mRNA, control gene expression in response to specific metabolites. We succeeded in re-engineering the first orthogonal riboswitches that are no longer triggered by the natural metabolites, but instead could be controlled by the addition of various synthetic heterocyclic molecules [5]. Our re-engineering approach provides a methodological platform for the creation of new orthogonal regulatory components and biosensors for synthetic biology and biotechnological applications including gene functional analysis, antimicrobial target validation and screening. For example, we have shown how mutually orthogonal synthetic riboswitches can be used to affect the simultaneous and independent control of multiple genes within the same cell [6]. In addition, we have demonstrated how the modular architecture of purine riboswitches can be exploited to develop orthogonal and chimeric switches that are transferable across diverse bacterial species, modulating either transcription or translation, to provide tunable activation or repression of target gene expression, in response to synthetic non-natural effector molecules [7]. Our novel riboswitch–ligand pairings are shown to regulate physiologically important genes required for bacterial motility in E. coli, as well as cell morphology in B. subtilis [7]. A rational targeted approach was also used to re-engineer the PreQ1 riboswitch from B. subtilis, into an orthogonal OFF-switch that can be controlled by the addition of a synthetic ligand [8].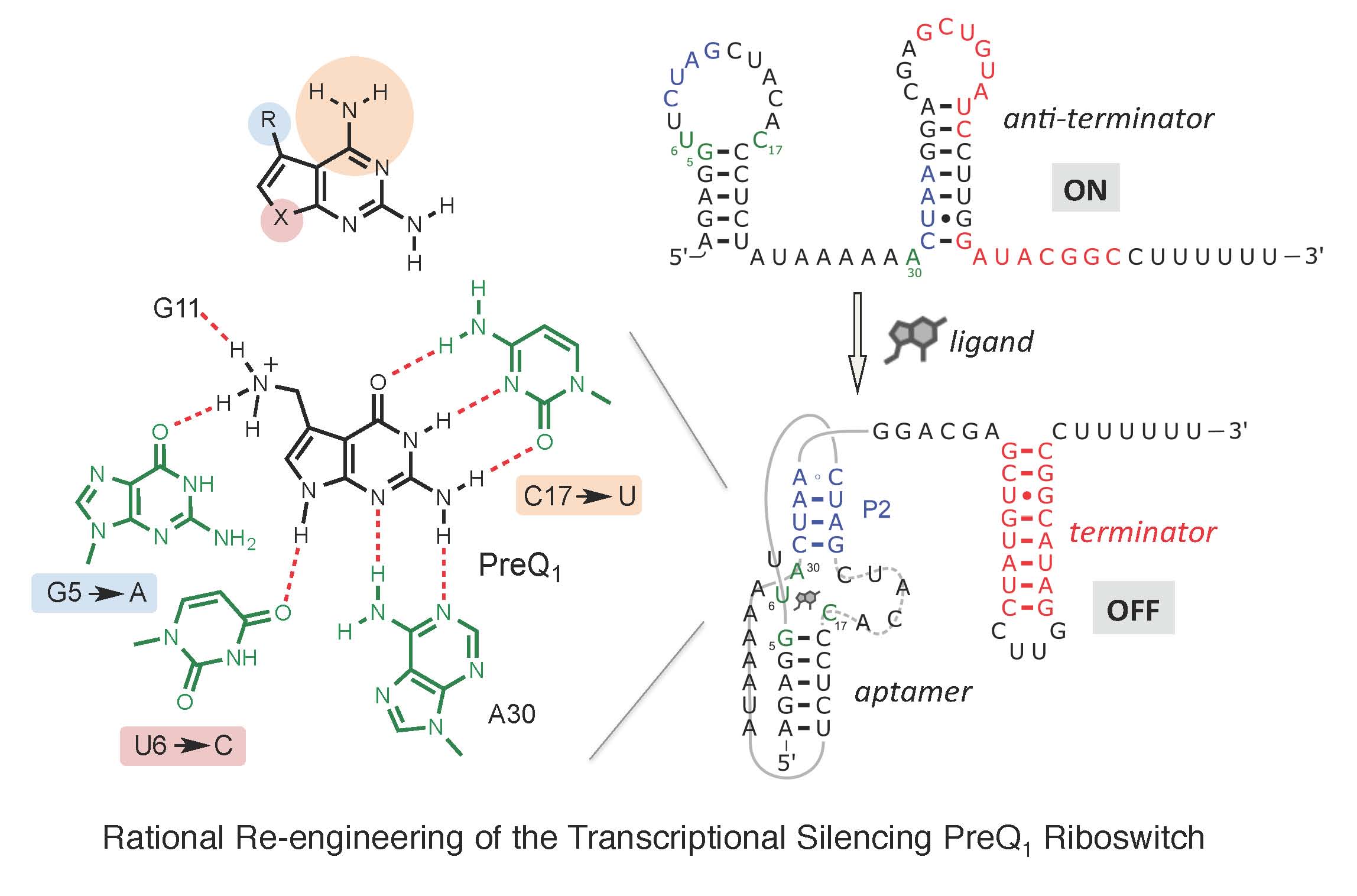
[1] K. J. Fettes, N. Howard, D. T. Hickman, S. A. Adah, M. R. Player, P. F. Torrence and J. Micklefield. Chem. Commun. 2000, 765-766; [2] D. T. Hickman, P. M. King, M. A. Cooper, J. M. Slater and J. Micklefield. Chem. Commun. 2000, 2251-225; [3] R. Worthington, J. Micklefield Chem. Eur. J.2011, 17, 14429-14441; [4] N. Bell, R. Wong, J. Micklefield Chem. Eur. J. 2010, 16, 2026-2030; [5] N. Dixon, J. Duncan, T. Geerlings, M. S. Dunstan, J. McCarthy, D. Leys & J. Micklefield. Proc. Natl. Acad. Sci. USA 2010, 107, 2830 [6] N Dixon, C Robinson, J. N. Duncan, T Geerlings, S. P. Drummond & J. Micklefield. Angew. Chem. Int. Ed. 2012, 51, 3620; [7] C. J. Robinson, H. A. Vincent, M.-C. Wu, P. T. Lowe, M. S. Dunstan, D. Leys, J. Micklefield. J. Am. Chem. Soc. 2014, 136, 10615; [8] M. C. Wu, P. T. Lowe, C. J. Robinson, H. A. Vincent, N. Dixon, J. Leigh & J. Micklefield. J. Am. Chem. Soc. 2015, 137, 901
